Dynamics of MREB filaments inside bacteria
Background: The cytoskeletal protein MreB is an essential component of the bacterial cell shape generation system. MreB interacts with cell wall synthesis machines, which produce peptidoglycan (PG) strands in hardly resolvable patterns. Inside living Bacillus subtilis cells MreB forms moving filamentous structures of variable lengths, typically not longer than one micrometer. Problem: What is the role of the MreB filaments in the context of cell wall synthesis and reorganization? We have made strange observations: These filaments move along their orientation and mainly perpendicular to the long bacterial axis revealing a maximal velocity at an intermediate length and a decreasing velocity with increasing filament length. Filaments move along straight trajectories, but can reverse or alter their direction of propagation. Approach: Approach: We use fast super-resolution microscopy based on TIRF-SIM to measure the strange dynamics of MreB filaments. We develop mechanistic and mathematical model consistent with all observations. In these models and observations the MreB filaments mechanically couple several motors that putatively synthesize the cell wall, whereas the filaments’ traces mirror the trajectories of the motors. We investigate whether the coupling of cell wall synthesis motors determines the MreB filament transport velocity, whereas the filament mechanically controls a concerted synthesis of parallel peptidoglycan strands to improve cell wall stability. |
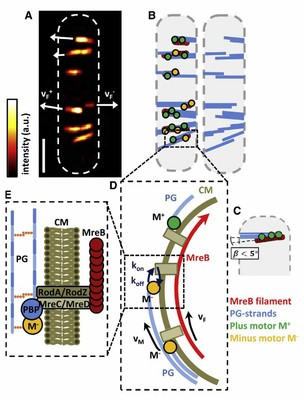
Schemes for MreB filament connection via membrane to motors as well as filament distribution. (A) TIRF-SIM image of MreB filaments and cell outline. Filament velocities (vF+, vF-) are indicated by white arrows. Scale bar is 1µm. (B) Left: corresponding possible distribution of p- and m-motors, M+(green) and M-(orange), as well as PG strands (blue). Resting filaments have an equal number of p- and m-motors attached. Right: the corresponding distribution of newly synthesized PG strands. (C) Scheme for an MreB filament organizing a concerted synthesis of three parallel PG strands by three synthesis motors. The angle ß<5° between the MreB filament and the PG strands cannot be resolved even with TIRF-SIM. (D) Motor movement enables filament transport with velocity vF in either the p or m direction as long as at least one motor is bound with rate kon via the transmembrane complex to MreB (koffis the unbindingrate). (E) A motor M is bound to the PG strands and is connected via a protein complex (PBPs, RodA/RodZ, and MreC/MreD) through the CM to an MreB filament
|
- Olshausen P, Soufo H, Wicker K, Heintzmann R, Graumann P, Rohrbach A
Superresolution Imaging of Dynamic MreB Filaments in B. subtilis - A Multiple-Motor-Driven Transport?
2013 Biophys J, Band: 105, Nummer: 5, Seiten: 1171 - 1181